Matthew Junker, Ph.D.
Associate Professor of Chemistry- Biochemistry
Contact Information
- Office: Boehm 314
- Phone: (610) 683-4199
- Email: junker@kutztown.edu
Courses Taught
- CHM 310, 312 Biochemistry I and II with laboratory
- CHM 214, 216 Organic Chemistry Laboratory I and II
- CHM 318 Advanced Biochemistry
- CHM 370 Research in Chemistry
- CHM 102 General Chemistry II
Research: Protein Structure and Function
Overview
Many cellular processes are carried out by proteins, tiny machines that catalyze chemical reactions, transport molecules within and between cells, provide cell architecture, and regulate gene expression and cell growth. The ability of proteins to carry out specific processes depends on their structure and energetics: active site geometries, complementation of recognition surfaces, conformational changes, affinities, the stability of folds. My research investigates the molecular mechanism of proteins that function in programmed cell death, gene expression, and metabolism. Almost all of the hands-on research is carried out by KU undergraduate students.
Experimental Approach
All of the research projects generally follow a common set of steps:
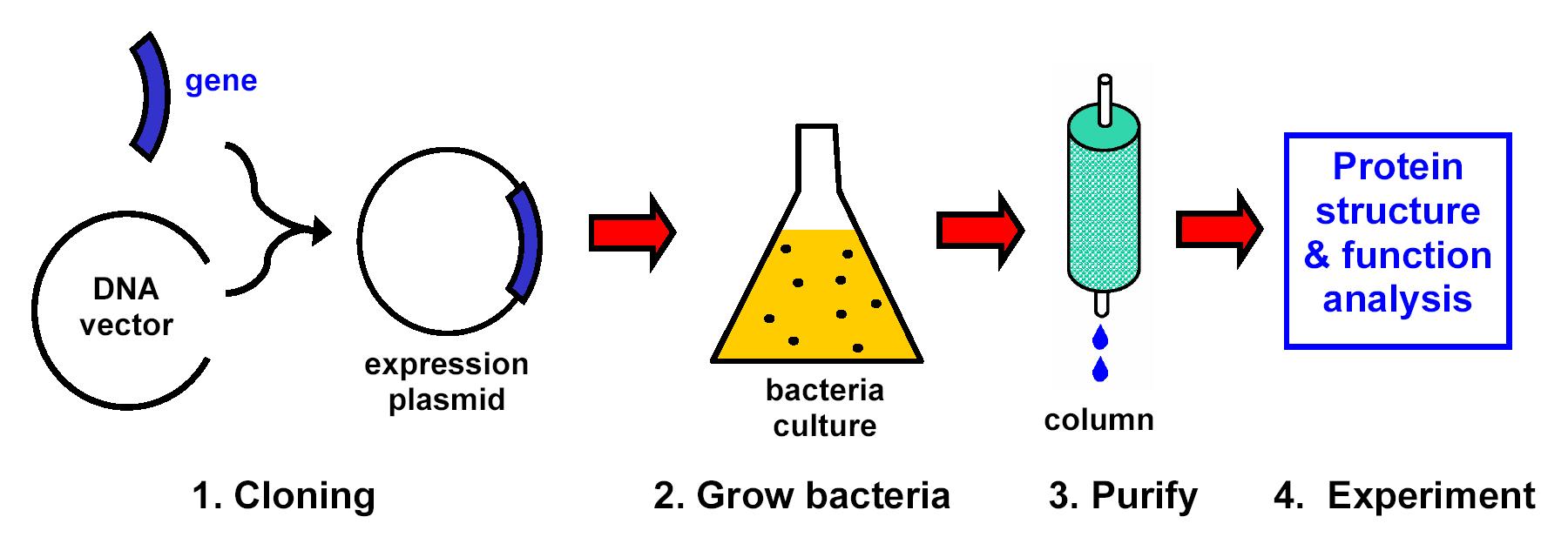
- Use recombinant DNA methods (cloning) to insert into bacteria the genes for proteins of interest.
- Grow bacteria to make the proteins of interest.
- Lyse (break open) the bacteria and purify the proteins of interest.
- Carry out complementary functional (biochemical assays) and structural studies of the purified proteins.
Example Research: Apoptosis
Apoptosis (programmed cell death) is a process in all animals that eliminates unneeded or potentially harmful cells. Dysfunction in apoptosis can lead to cancer or neurodegenerative diseases. Within cells, apoptosis is controlled by proteins- large complex molecules whose function depends on their correct folding and structure. Apoptosis requires protein enzymes called caspases. Caspases are kept inhibited in living cells by an inhibitor of apoptosis proteins (IAPs). Apoptosis occurs when certain stimulator proteins bind to IAPs to de-inhibit (activate) caspases.
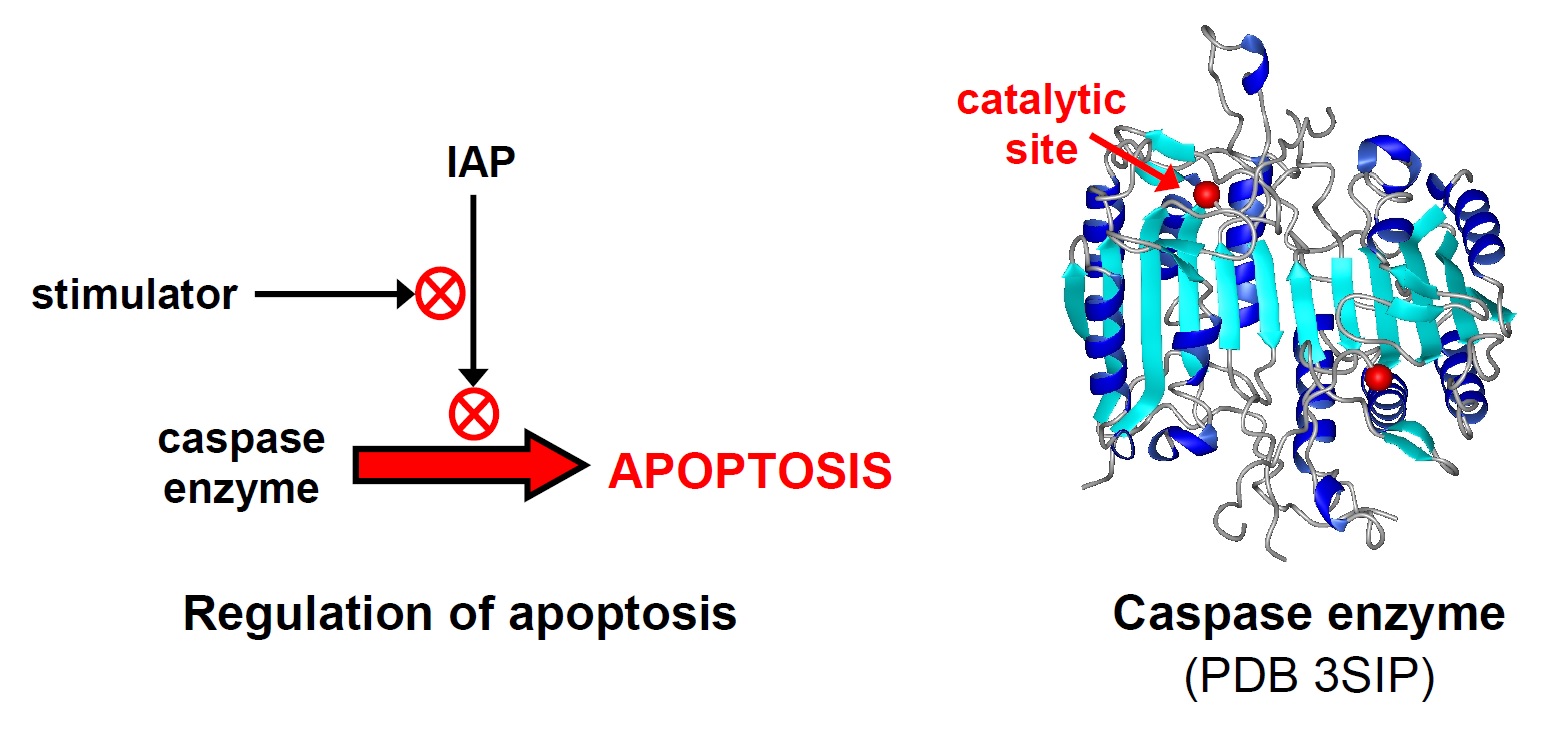
We are investigating how apoptosis proteins function at the level of their molecular structure and chemistry. We study human and Drosophila (fruit fly) proteins, using recombinant DNA methods to modify the proteins and synthesize them in large amounts in bacteria. One area of focus is determining how different regions of IAPs contribute to regulating caspase activity.
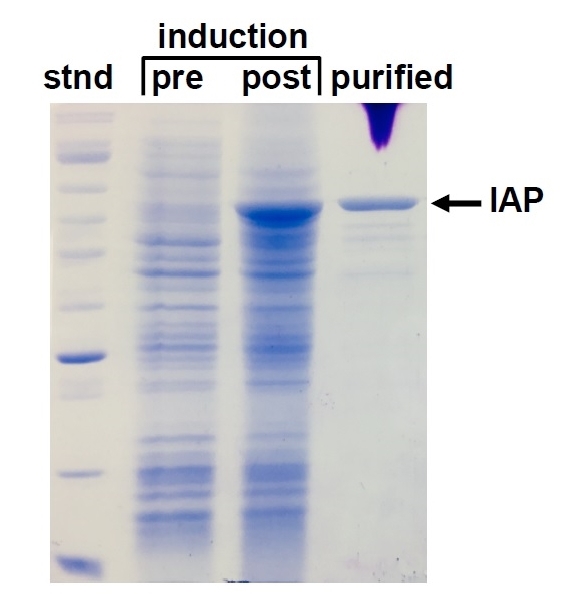
This SDS-PAGE gel shows the synthesis and purification of a Drosophila IAP produced in E. coli. The pre- and post-induction samples show all E. coli proteins before and after turning on expression of the IAP gene. The final lane shows the IAP protein after purification. "Stnd" is a sample of protein standards for size reference.
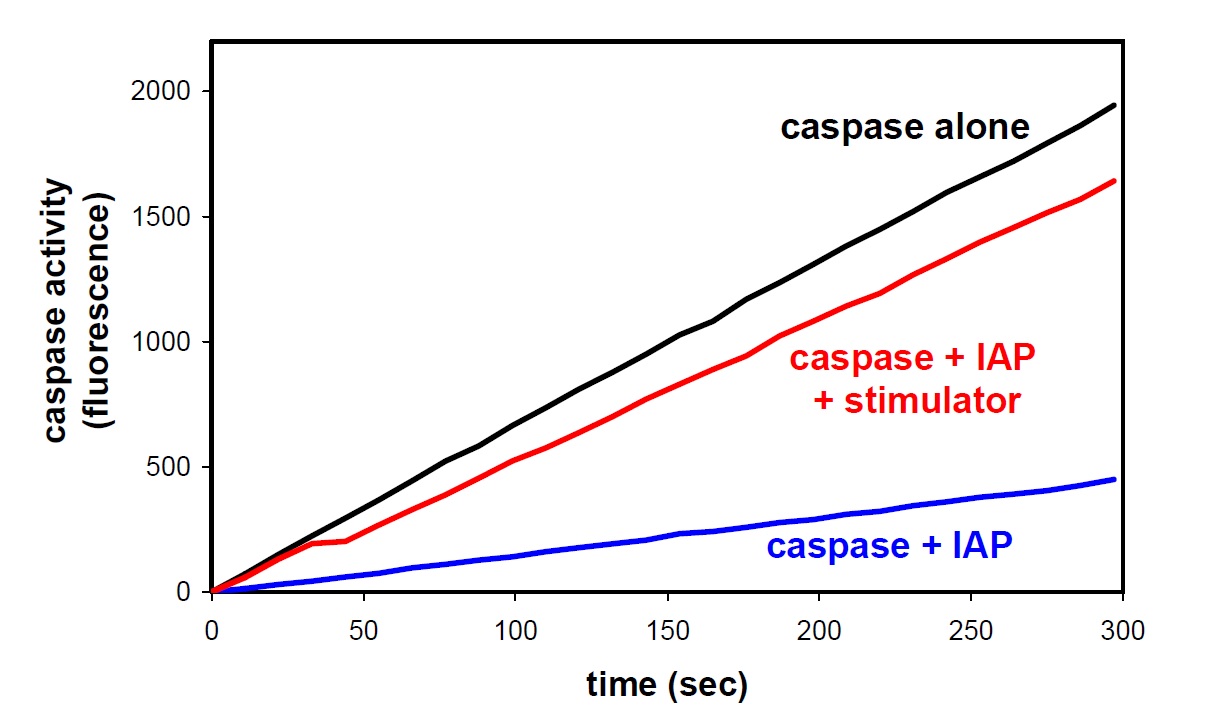
Caspase function is measured using a fluorescence-based caspase assay in which caspase reacts with a substrate to produce a fluorescent product. The assay shows how IAPs inhibit caspase function and stimulators reverse this inhibition, all with purified proteins.
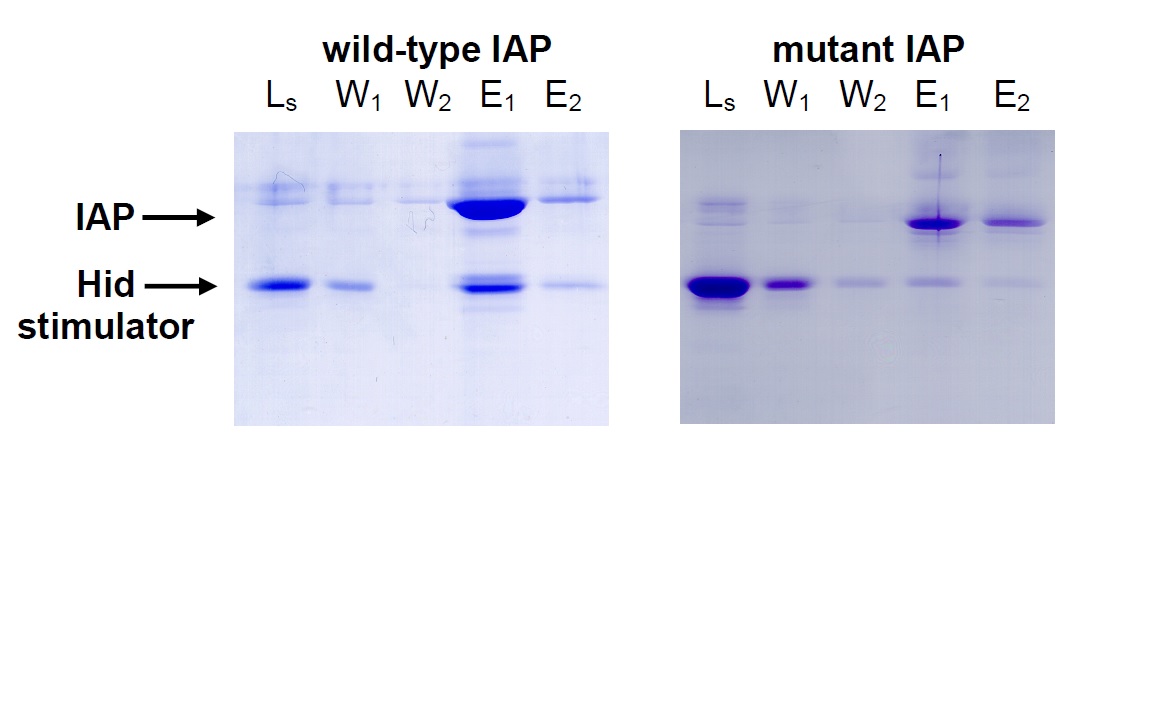
Cells with a mutated IAP had been reported to be resistant to apoptosis induction. Using an IAP-binding column, we determined that the mutant IAP could no longer bind and co-elute stimulators (L= load Hid, W= wash, E= elute).
Other Current Research Topics
Other ongoing research projects include developing a new method to control protein self-association and, in collaboration with Dr. Carsten Sanders in the KU Chemistry & Biochemistry Program, investigating the mechanism for the enzyme cytochrome c heme lyase (CCHL). The projects all involve the same basic technologies of generating purified proteins by recombinant DNA methods and assaying their function and structure in biochemical experiments.
Recent Conference Presentations
* denotes undergraduate students
S. Shara* and M. Junker (2022) A New Method to Self-Associate Proteins Using a Cooperative DNA-Binding Protein. ASBMB (American Society for Biochemistry and Molecular Biology) Annual Meeting, Philadelphia, PA, April 2022.
T. Hawes*, C. Emerich*, and M. Junker (2020) Self-association enhances the potency of an inhibitor of apoptosis protein. American Chemical Society National Meeting and Expo, Philadelphia, PA, March 2020 (virtual due to COVID).
B. Graver*, M. Storm*, and M. Junker (2019) The mechanism of inhibitor of apoptosis protein (IAP) domain binding to an apoptotic caspase. Middle Atlantic Regional Meeting of the American Chemical Society, Baltimore, MD, May 2019.
R. Roberts*, M. Junker, and C. Sanders (2019) The involvement of amino acid residues E133 and R199 in the heme-to-apocytochrome c ligation mechanism of cytochrome c heme lyase (CCHL). Thomas Jefferson University Sigma Xi Research Day, April 2019.
M. Storm* and M. Junker (2018) Dissecting the function of IAP (Inhibitor of Apoptosis) protein domains in inhibiting an apoptotic caspase. ASBMB (American Society for Biochemistry and Molecular Biology) Annual Meeting, San Diego, CA, April 2018.






